DNA sequencing has emerged as a key component of microbial analysis due to the explosion of studies on the human microbiome. 16S rRNA gene sequencing and shotgun metagenomic sequencing are two of the most powerful methods for profiling microbial populations, especially when labs use 16s metagenomic service for accurate microbial detection.
Although they both aid in the identification of microorganisms from complex samples, their goals, methods, resolutions, and results are very different. For researchers, physicians, and public health specialists alike, it is essential to comprehend these distinctions, especially when evaluating the advantages of 16s rrna sequencing in comparison to whole-genome approaches.
So, here in this blog, we have shared everything that you need to know about the 16S rRNA and metagenomic sequencing.
Continue reading to learn more.
Table of Contents
ToggleWhat Are 16S rRNA and Metagenomic Sequencing?
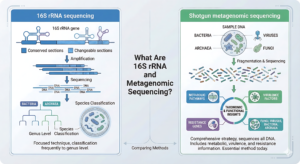
16S rRNA sequencing
The 16S ribosomal RNA gene, a conserved genetic marker found in all bacteria and archaea, is amplified and sequenced using the focused technique known as 16S rRNA sequencing. Both highly conserved and changeable sections of the 16S gene enable the classification of species, frequently down to the genus level.
Shotgun metagenomic sequencing
Shotgun metagenomic sequencing adopts a more comprehensive strategy. It sequences all of the DNA in a sample, including those of viruses, fungi, bacteria, and archaea. It provides taxonomic and functional insights that also include information on metabolic pathways, virulence factors and resistance genes, making it essential among metagenomic sequencing methods today.
How does each method work?
16S rRNA workflow
- Primer selection & PCR amplification
Universal primers are used to target conserved areas that surround variable portions of the 16S gene. These primers use the polymerase chain reaction (PCR) to amplify the 16S gene’s target regions.
- Library preparation
Cleaners and adapters are used to get amplified DNA ready for sequencing.
- Sequencing
The amplicons are read by high-throughput sequencers
- Bioinformatics analysis
For taxonomy identification, tools such as QIIME2 or Mothur compare reads to databases like SILVA or RDP.
Metagenomic sequencing workflow
- DNA Extraction
All of the sample’s genomic DNA, including those of bacteria, viruses, fungi, and hosts, is removed, enabling deeper organism profiling through metagenomic dna sequencing.
- Library preparation
DNA is randomly broken apart and ligated using sequencing adapters to prepare the library.
- Sequencing
Shotgun sequencing is carried out using technologies
- Assembly and annotation
Using programs like MetaPhlAn, HUMAnN, or MG-RAST, sequencing reads are put together into contigs and annotated.
Key Differences
Taxonomic depth:
Microbial communities are usually resolved at the genus level using 16S rRNA. Although full-length 16S sequencing and high-quality reads can enable species-level identification, sequence similarity frequently places restrictions on this process.
Deeper taxonomic resolution, frequently down to the species or strain level, is provided via metagenomics. Additionally, it can identify new or unidentified bacteria that 16S surveys missed.
Functional insights:
Only offers taxonomic information. It is unable to identify functional traits like virulence factors, resistance genes, or metabolic potential.
Beyond taxonomy, metagenomics reveals metabolic pathways, antimicrobial resistance (AMR) genes, and functional genes. In clinical and environmental studies, where functioning is important, this is crucial.
Comparative Pros & Cons
Accuracy, bias, and contamination limitations
16S rRNA:
- Pros: Bacterial identification is sensitive and specific.
- Cons: Results may be skewed by chimera formation, primer mismatches, and PCR biases.
Shotgun Metagenomics:
- Pros: Complete, unbiased profiling.
- Cons: Contaminant DNA, particularly host DNA in clinical samples, may have an impact. needs a lot of filtration and disinfection.
Cost, time, and data volume
- 16S rRNA: Perfect for large-scale screening, it is quicker and more economical with smaller datasets.
- Metagenomics: Time-consuming and costly, but its application in high-resolution research is justified by the volume and variety of data it provides.
Bioinformatics complexity
- 16S rRNA: Bioinformatics pipelines that are simpler. Analysis is available thanks to user-friendly technologies.
- Metagenomics: Needs sophisticated knowledge of bioinformatics. Processing big and complicated datasets requires a strong computational infrastructure.
Which Method to Use? Application-Based Comparison
Clinical diagnostics:
- 16S rRNA: Rapid detection of microorganisms in illnesses such as pneumonia or sepsis. restricted to infections that are unknown or polymicrobial.
- Metagenomics: Identifies novel pathogens, mixed infections, and AMR genes; particularly helpful when culture findings are negative.
Environmental studies:
- 16S rRNA: Assessing biodiversity in soil, water, or air microbiomes is frequently done using 16S rRNA.
- Metagenomics: Shows metabolic potential and diversity, including pollution degradation, carbon cycling, and nitrogen fixation.
Food safety:
- 16S rRNA: Checks for typical microbiological pollutants in food preparation and storage.
- Metagenomics: Particularly in intricate supply networks, metagenomics monitors new infections, resistance genes, and spoiling microbes.
Microbiome discovery:
- 16S rRNA: Excellent for preliminary mapping of the oral, cutaneous, or gut microbiomes is 16S rRNA.
- Metagenomics: Understanding disease correlations, host-microbe interactions, and potential treatment targets all depend on metagenomics.
Content Gap Insertions
Database integration:
- Curated taxonomy databases like SILVA, Greengenes, and RDP are essential for 16S rRNA.
- Shotgun metagenomics makes use of more extensive databases like KEGG, CARD, and RefSeq for functional annotation and MetaPhlAn for taxonomy.
Emerging hybrid approaches:
- More recent sequencing techniques improve species- and strain-level resolution by combining full-length 16S sequencing with long-read platforms such as Uncoded
- To take advantage of taxonomic clarity and functional insights, some researchers employ hybrid pipelines that integrate 16S and metagenomics.
Quality Control & Troubleshooting Tips:
- Incorporate both positive and negative controls into each run.
- Use strict filtration and disinfection procedures.
- To verify pipeline performance, use simulated communities.
Case Study Examples
16S rRNA Sequencing:
In a study on inflammatory bowel disease (IBD), 16S sequencing was used to find that patients’ gut microbiota had changed Firmicutes/Bacteroidetes ratios and decreased microbial diversity.
Metagenomics Sequencing:
In a neonatal intensive care unit outbreak, metagenomic sequencing assisted in locating a multi-drug resistant strain of Klebsiella that was missed by conventional culture techniques. Decisions about infection control were also informed by the discovery of mobile genetic elements and resistance genes.
Future Outlook & Trends
Long-read sequencing:
Complete microbial genome reconstruction from complex samples is now possible thanks to developments in long-read sequencing . In metagenomic research, this enhances taxonomic resolution and assembly precision.
Multi-omics integration:
Multi-omics, which combines metagenomics with proteomics, metabolomics, and metatranscriptomics, is the direction of future study. Microbial ecosystems and their interactions with hosts or environments will become more dynamic and systems-level as a result.
Conclusion
Shotgun metagenomic sequencing and 16S rRNA sequencing have both transformed our understanding of microbial communities, but they have different uses. 16S rRNA is an excellent option if you’re searching for bacteria in a sample and is also a cost-effective method.
On the other hand, shotgun metagenomics is a preferable option if your research requires more in-depth information, such as identifying bacteria down to the strain level or comprehending their roles, resistance to antibiotics, or metabolic pathways.
If you’re comparing 16S sequencing vs metagenomics, you’re not alone.
- Choose 16S rRNA sequencing if you want a simpler, cost-effective way to identify bacteria in your sample.
- Go for metagenomic sequencing if you need deeper insights, like strain-level detection or antibiotic resistance.
At Uncoded, we offer both. Our 16S V3–V4 Library Preparation Kit is trusted for accurate and easy microbial profiling.
And if your study needs more depth, our shotgun metagenomic solutions are ready to support you.
Still confused? Explore the kits, compare your options, and choose what fits your research best, whether you’re in a classroom or a clinical lab

