16S V3-V4 Library Preparation Kit


Introducing the 16S V3–V4 Library Preparation Kit for Metagenomics
Metagenomics involves examining genetic material obtained directly from diverse samples, allowing researchers to investigate microbial diversity without the necessity for culturing. This potent method is vital for comprehending microbial communities in various settings—including human health, agriculture, clinical diagnostics, and environmental monitoring. As microbiome research continues to grow, metagenomics has become indispensable across various fields, spanning human health, biotechnology, and environmental science.
Uncoded proudly introduces its product—the 16S V3–V4 Library Preparation Kit for metagenomic sequencing. This kit enables scientists to investigate microbial communities by focusing on the 16S ribosomal RNA (rRNA) gene, a well-conserved marker found in both bacteria and archaea. Its integrated hypervariable regions facilitate precise identification of various bacterial taxa, making it perfect for extensive microbial profiling.
What is the 16S V3–V4 Region?
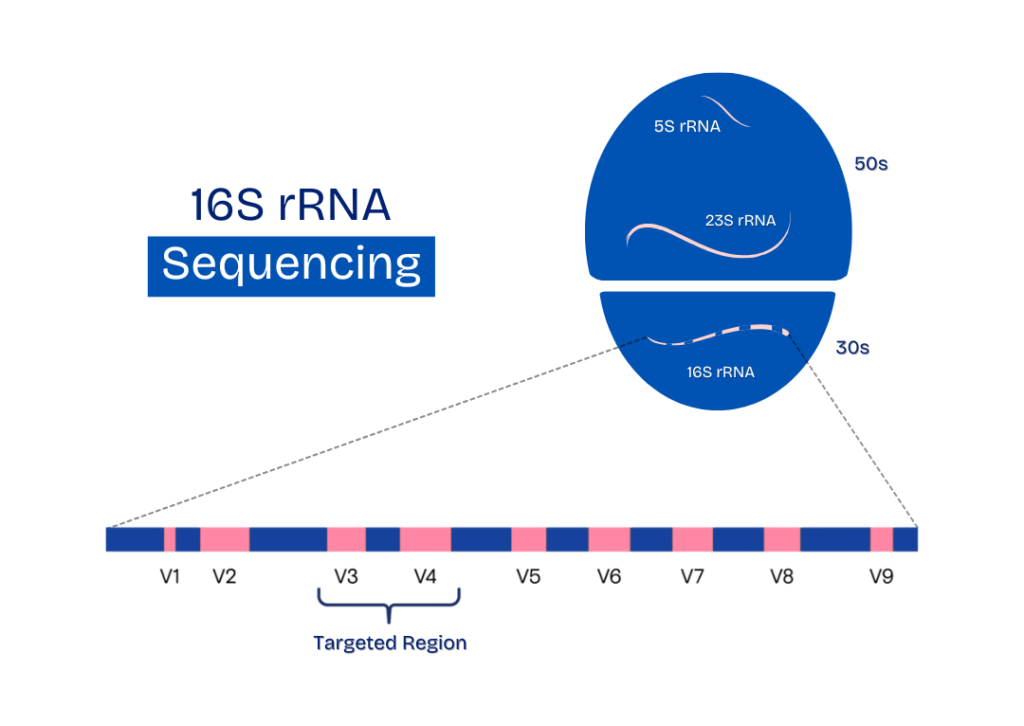
The 16S rRNA gene is a highly conserved genetic marker present in all prokaryotic organisms, which makes it an excellent target for identifying bacteria and conducting phylogenetic analyses. This gene contains nine hypervariable regions (V1–V9), each providing varying degrees of taxonomic resolution.
The V3–V4 region, which is roughly 460 base pairs long, is among the most informative and commonly utilized segments for microbial profiling. It offers a practical compromise between amplicon length and classification precision, making it particularly suitable for use with Illumina sequencing technologies.
Why 16S rRNA Gene Sequencing and the V3–V4 Region Matter
The 16S rRNA gene sequencing, initially introduced by Carl Woese and George E. Fox in 1977, has established itself as the definitive method in the fields of microbial taxonomy and ecology. The unique combination of conserved and variable regions within the gene allows for widespread detection as well as accurate differentiation between bacterial taxa.
Focusing on the V3–V4 region improves the resolution of intricate microbial communities, enabling researchers to differentiate between closely related species while ensuring dependable amplification. This region has strong support from comprehensive reference databases, establishing it as a reliable and reproducible option for metagenomic research across various sample types.

Addressing Major Challenges in Metagenomic Sequencing
Although metagenomics research is gaining popularity, it frequently encounters practical issues such as lengthy procedures, high expenses, variable sequencing quality, and limited detection sensitivity for samples with low biomass.
The 16S V3–V4 Library Preparation Kit from Uncoded tackles these challenges with several innovative features:
• Phased primers to minimize amplification bias and enhance coverage
• High-quality reagents to guarantee consistency and precision
• A simplified, user-friendly protocol that makes library preparation easier
• Wide compatibility with challenging sample types like stool and soil
• Optimized for a broad range of bacterial loads, ensuring accurate detection from low to high microbial concentrations
This comprehensive approach makes the kit a dependable choice for researchers working with complex or degraded samples, improving reproducibility and minimizing data loss.
Differential Uses in Industry and Research
The purpose of the 16S V3–V4 Library Preparation Kit is to facilitate a broad range of industrial and research applications by giving users practical knowledge about microbial communities in many sectors:
• Microbiome Research
In-depth characterization of microbial populations in human-associated areas, such as the gut, skin, and oral cavity, is made possible by microbiome research, which provides information on host microbe interactions, health, and illness.
• Microbiology in Clinical Practice
Helps with infection surveillance, diagnosis, and research on antibiotic resistance by making it easier to identify harmful and beneficial bacteria in clinical samples.
• Food Microbiology
To ensure food safety, traceability, and quality control, food microbiology helps monitor the microbial load, spoilage organisms, and foodborne pathogens in raw materials and processed foods.
• Soil and Agricultural Microbiology
Aids in the monitoring of plant-associated microbiota, the assessment of soil health, and the investigation of microbial interactions that affect nutrient cycle, crop yield, and insect resistance.
• Environmental Surveillance
By identifying microbial populations in water, air, and waste systems—critical for ecosystem monitoring and remediation efforts—it supports pollution studies and biodiversity assessments.
• Applications in Industry and Biotechnology
The characterization of microbial consortia involved in industrial workflows improves process optimization in the fields of fermentation, bioproduction, and microbial enzyme research.
• Biofuels and Energy
Promotes sustainable energy solutions by making it possible to investigate the microbial populations involved in the generation of bioenergy, such as those employed in anaerobic digestion, bioethanol, and biogas systems.

Applications of 16S V3-V4 Library Preparation Kit
Consistency
Ensures reliable, reproducible results across different samples without background noise by optimized protocol and implementing QMS guidelines
User-Friendly
Kits intuitive design, allowing for easy & straightforward workflow with minimal hands-ontime & reduce technical complexity
Scalability
Effectively supporting both small & large scale projects with consistent performance & adaptability across various sample types for different Illumina platforms
Technical Support
Benefit from our dedicated technical support team from expert guidance and troubleshooting
| Cat. No. | Rxns |
|---|---|
| UN16S/0507/024 | 24 |
| UN16S/0507/096 | 96 |
| UN16S/0507/384 | 384 |

| Storage temperature: -25 °C to -15 °C | |
| Ref No: UN16S/0507/096/100 | 16S V3-V4 Library Preparation kit (1 of 3) |
| Storage temperature: 2°C to 8°C | |
| Ref No: UN16S/0507/096/200 | 16S V3-V4 Library Preparation kit (2 of 3) |
| Storage temperature: -25 °C to -15 °C | |
| Ref No: UN16S/0507/096/300 | 16S V3-V4 Library Preparation kit (3 of 3) |

Frequently Asked Questions
Curious about workflow, accuracy, or compatibility? Find all your answers here
High-quality genomic DNA with optimal DNA purity is essential for successful 16S metagenomics analysis. One key measure of purity is the 260/280 ratio, which should ideally be between 1.8 and 2.0 to indicate minimal contamination. The DNA should be free from contaminants like heme, humic acid, polysaccharides, polyphenols, or any substances carried over from the sample source. Contaminants introduced during DNA extraction can impact the quality of the sequencing run, so it is important to ensure that the DNA is pure and clean to achieve reliable results.
Use the Qubit (fluorescence-based method), as it provides more accurate quantification of DNA. Although Nanodrop (absorbance-based) can offer useful information on sample purity, it is not sufficient for accurate quantification.
The standard protocol for 16S V3-V4 Library preparation kit requires between 1-5 ng of total DNA (high quality). For this kit, 12.5 ng is recommended, and sufficient yields are attainable from as little as 100pg-50ng depending on the sample type.
16S V3-V4 Library preparation workflow includes library preparation, sequencing on the Illumina® platform and data analysis.
Our targeted 16S-library preparation kit typically provides sensitivity down to the genus level.
The 16S V3-V4 Library Preparation offers several advantages over others, including industry-leading turnaround time. It is based on the phased primer concept to specifically target the highly variable V3 and V4 regions of the 16S rRNA gene, which enhances sensitivity and accuracy. The assay is cost-effective and has been validated across various sample types, ensuring reliable performance in diverse applications.
What are the challenges of 16S V3-V4?
How does16S V3-V4 improve compliance?
Lorem ipsum dolor sit amet consectetur. Dolor aenean lorem integer nunc at nascetur enim. At porta eu imperdiet porta condimentum vitae in lacus. Amet placerat senectus justo integer nulla lobortis bibendum sed quam. Morbi consequat iaculis et est elementum in.
What is 16S V3-V4?
Lorem ipsum dolor sit amet consectetur. Dolor aenean lorem integer nunc at nascetur enim. At porta eu imperdiet porta condimentum vitae in lacus. Amet placerat senectus justo integer nulla lobortis bibendum sed quam. Morbi consequat iaculis et est elementum in.
How does16S V3-V4 improve compliance?
Lorem ipsum dolor sit amet consectetur. Dolor aenean lorem integer nunc at nascetur enim. At porta eu imperdiet porta condimentum vitae in lacus. Amet placerat senectus justo integer nulla lobortis bibendum sed quam. Morbi consequat iaculis et est elementum in.
Learn more about : 16S V3-V4 Library Preparation Kit