16S metagenomics is a powerful tool used to perform microbial diversity analysis and understand the microbial diversity within environmental and biological samples. It focuses on sequencing and analysing the 16S ribosomal RNA (rRNA) gene, a critical component of the bacterial ribosome, which is highly conserved across different bacterial species but contains variable regions that can provide unique genetic signatures for bacterial identification. This process is central to what is 16s metagenomic sequencing.
The 16S rRNA gene is part of the small subunit of the bacterial ribosome and is crucial for protein synthesis. It is approximately 1,500 base pairs long and contains both conserved and variable regions:
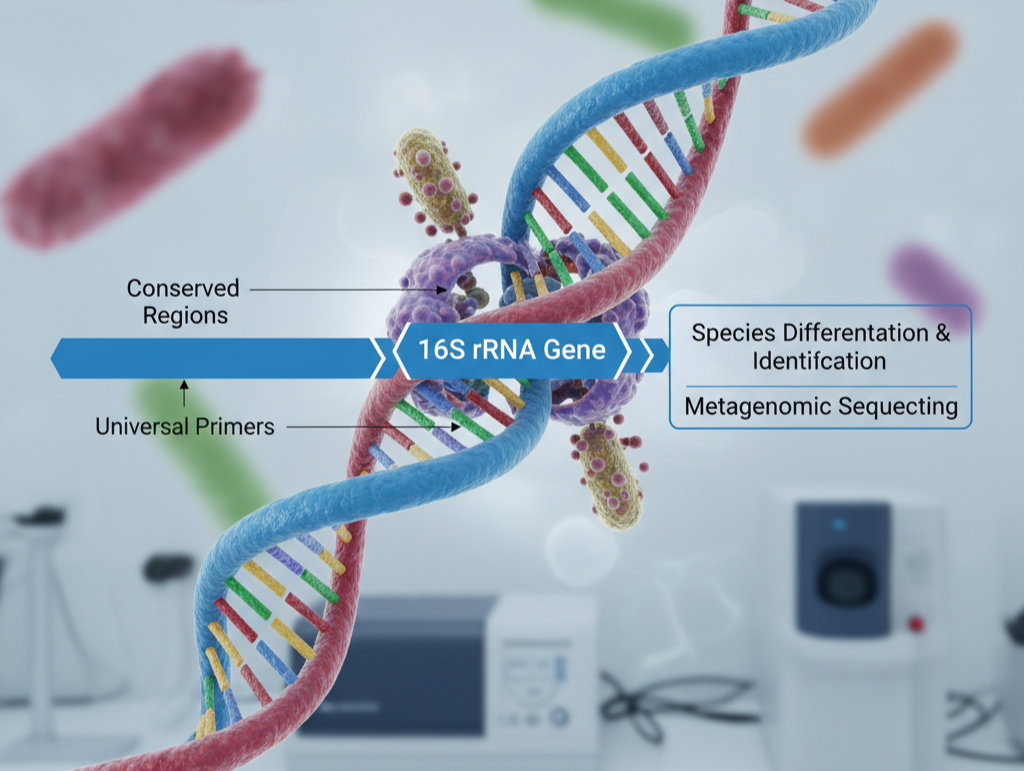
- Conserved Regions: These are similar across most bacterial species and are useful for designing universal primers for PCR amplification.
- Variable Regions: These regions differ between bacterial species and are used to differentiate and identify them. These regions are crucial when building a 16s metagenomic sequencing library for analysis.
The process of 16S metagenomics typically involves several key steps:

- Sample Collection: Environmental or biological samples (such as soil, water, gut microbiota) are collected for analysis.
- DNA Extraction: Total DNA is extracted from the sample, which includes the DNA of all microorganisms present.
- PCR Amplification: Specific primers are used to amplify the 16S rRNA gene from the extracted DNA.
- Sequencing: The amplified 16S rRNA gene is sequenced using high-throughput sequencing technologies (e.g., Illumina, Ion Torrent), a method also referred to as 16s rrna sequencing.
- Data Analysis: Sequencing data are processed to identify bacterial taxa by comparing the variable regions of the 16S rRNA gene to a reference database. This approach is a cornerstone of 16s metagenomic sequencing.
16S metagenomics has a wide range of applications, including:

- Microbial Diversity Studies: Provides insights into the diversity and composition of microbial communities in various environments.
- Human Health: Helps in understanding the human microbiome and its role in health and disease, such as gut microbiota analysis in conditions like obesity, diabetes, or inflammatory bowel disease.
- Environmental Monitoring: Used to study microbial communities in different ecosystems, including soil, water, and extreme environments.
- Biotechnology: Assists in discovering new microbial strains with potential industrial applications, such as in bioremediation or fermentation.
16S Metagenomics offers great advantages when analysing biological samples by providing-
-
- High Resolution: Provides detailed information about microbial community composition.
- Non-Invasive: Suitable for a variety of sample types, including complex environments and human samples. Using a 16s metagenomic sequencing library ensures standardized and reproducible results.
Conclusion:
16S amplicon sequencing is a valuable technique for studying microbial communities and understanding their roles in various contexts. By leveraging the 16S rRNA gene’s unique features, researchers can gain insights into microbial diversity, functional potential, and interactions in complex environments. The 16S amplicon sequencing and metagenomic sequencing methodologies both have extensive applications. It can also be employed to investigate the diversity and composition of microbial communities in varied environments such as soil, water bodies, and animal intestines.

